DNA.I.
Early findings and emerging questions on the use of AI in genomics
31 August 2023
Reading time: 70 minutes
Executive summary
In recent years, the scientific fields of artificial intelligence (AI) and genomics have experienced increased public attention and investment by public and private institutions. The UK Government, for example, has made explicit plans to become ‘the most advanced genomic healthcare system in the world’, and lists AI as one of five ‘critical technologies’ that can make the country a scientific superpower.[1]
Both AI and genomics have already been used to address major scientific challenges, including genomic sequencing to identify novel COVID-19 variants[2] and the use of AI and machine learning to predict the structure of proteins.[3] But both fields have also resulted in controversies over their ethical and societal implications, and raised a host of difficult issues for those looking to regulate, direct and govern their development and use. In genomics, recent debates about acceptable uses of CRISPR-Cas9 have raised concerns around the ethics of genetic engineering.[4] Similarly, the field of AI has recently experienced an increasingly intense public conversation about the ethical and societal implications of foundation models, powerful AI systems capable of a wide range of general tasks.[5]
AI and genomics are also becoming progressively more intertwined. Many recent advances in genomics have been made possible by the use of AI,[6] and AI research and product teams have increasingly sought to use genomic data to create AI-powered genomics research and products.[7] Economic forecasts have suggested the market for AI and genomics technologies could reach more than £19.5 billion by 2030, up from half a billion in 2021.[8]
The increasing convergence of AI and genomics is set to present policymakers with a new set of practical and theoretical challenges. Considered separately, developments in AI and in genomics already pose deep questions concerning agency, privacy, quality, bias and power. Considered in relation to one another, the issues posed by the two technologies become harder to predict, more complex and more numerous.
While there has been much research considering the ethical impacts of AI and genomics as separate technologies, comparatively little attention has been paid to exploring the broader implications of the two technologies when used together, and from a structural perspective. For policymakers seeking to navigate and regulate AI and genomics, this is a critical evidence gap.
AI and genomics futures is a joint project between the Ada Lovelace Institute and the Nuffield Council on Bioethics that investigates the ethical, and political economy issues arising from the application of AI to genomics – which we refer to throughout this report as AI-powered genomics.
This report of our early findings sets out the results of our research, its significance for policymakers, and the specific topics and questions we will focus on.
Our research shows that:
- AI-powered genomics has seen significant growth in the past decade, driven principally by advances in machine learning and deep learning, and has developed into a distinctive, specialised field.
- Private-sector investment in companies working on AI-powered genomics has been substantial – and has mainly gone to companies working on data collection, drug discovery and precision medicine.
- The most prominent current and emerging themes in research on AI-powered genomics relate to proteins and drug development, and the prediction of phenotypic traits from genomic data.
Moreover:
- The specific combination of emerging themes and capabilities identified in AI-powered genomics points to the increasing viability of two broad techniques within healthcare over the next five to ten years:
- AI-powered genomic health personalisation: the ability to understand how treatment for the same health condition might vary between different people on the basis of genomic variations, and to tailor and adapt treatments accordingly.
- AI-powered genomic health prediction: the use of genomic data to estimate the probability of different people developing particular health conditions, responding well or badly to particular medicines or treatments, or being affected by lifestyle factors.
- The potential emergence of these techniques raises profound, urgent ethical, legal and policy questions.
- While some of these issues are already discussed and accounted for in existing legal, ethical and policy discourse, there are many questions concerning the macro-level impacts of developments in AI-powered genomics that have yet to be adequately explored.
- In particular, there is an urgent, relatively unmet need for sustained thinking and research on the structural, political, and economic implications of AI-powered genomic health prediction, and how its development might be steered and governed in line with public values and priorities.
How to read this report
This report sets out early findings and future focus of the AI and genomics futures project, as well as explaining its more specific research aims and methodologies. While the report will make most sense read in order, its three chapters can also be read alone.
If you are short of time and mainly interested in:
- the research methods deployed by the project, read the chapter on ‘Scope, research questions and methodology’
- the findings from our literature review, scientometric analysis and horizon-scanning exercise, read the chapter on ‘Detailed research findings’
- why the societal implications of the application of AI and genomics deserve the urgent attention of policymakers, and what areas we think are especially in need of exploration, read the introduction and the chapter on ‘Key research findings and their implications’.
Introduction
The past decade has seen a surge of research and investment (from commercial and government sources) into the use of AI to advance genomic science.
This increase in activity is largely due to the potential of advances in machine learning and deep learning to yield substantial improvements in the collection, analysis and useful deployment of genomic data.[9] As a result, there has been a steady rise in academic interest in the topic. Growth in the number of scientific papers published on AI and genomics has been accelerating year on year since 2017.[10] There have also been buoyant and wide-ranging predictions regarding the growth of the market for AI in genomics over the next decade.[11]
The impacts of such developments could be considerable. Potential implications include a paradigm shift in drug development and the ability to better predict complex human traits (such as height, body mass index or diabetes risk) on the basis of genomic data.
But the technical and societal implications of AI-powered genomics are by no means straightforward or certain. Among experts, accounts differ regarding the speed, extent and significance of the transformations promised – and the degree to which AI will help overcome challenges faced by traditional genomic science.[12] [13] Likewise, both AI and genomics are scientific fields that have prompted major concerns around their implications for society, including questions around bias and discrimination,[14] worries around corporate capture,[15] and issues relating to privacy and the use of data.[16] As a combination of two of the most controversial technologies of the 21st century, research, the development and deployment of AI-powered genomics will present considerable ethical, political and legal challenges.
For practitioners and decision-makers concerned with how we cultivate, manage and regulate genomics, AI-powered genomic science raises a host of difficult and important questions. Where, how and to what extent is AI currently changing the capabilities, viable applications and practice of genomic science? What future changes are anticipated – and how confident can we be of their emergence? What might be the political, economic and societal impacts of these changes? And, critically, what should we do now, in light of these possibilities, to ensure that AI-powered genomics develop and are deployed for the public good and in accordance with societal values?
We don’t yet have good answers to these questions. Though there is a large and growing discourse on the societal impacts of AI and genomics – separately – there is far less guidance on the effects these two fields might have in combination. There is also less work on the implications of the most imminent advances in AI-powered genomics for how different groups and actors within society relate to and behave towards one another. From the perspective of a policymaker thinking about the governance of genomic science in the face of AI-powered developments, there is a particular need for:
- Analysis of the opportunities and challenges posed by AI and genomics when used together.
If many of the most significant developments in genomics are set to be driven by AI, then there also needs to be explicit analysis of the ethical issues that might be encountered when these two technologies are combined. These issues may differ in both kind and degree from those with which we are already familiar. - An analysis of who and what is driving advances in AI-powered genomics.
The past few years have seen a growing body of research interrogating the power dynamics and incentive structures created by AI (partly in response to charges of ‘ethics washing’ and a proliferation of ethical principles for AI whose application to concrete problems is often unclear).[17] There is less work on the potential political economy of genomic science, however. Identifying the major actors driving these advances, what their incentives are, and intended beneficiaries will help policymakers, funders, and developers to better understand and address possible long-term impacts of AI-powered genomics. - A clear assessment of the predicted significance and imminence of AI-powered developments in genomic science.
To develop an effective policy response to the rise of AI-powered genomics, it will be necessary to have a clear understanding of which aspects of the technologies are likely to be most impactful, which are (in the absence of government intervention) likely to develop fastest, and which will diffuse throughout society first.
The AI and genomics futures project, jointly conducted between the Nuffield Council on Bioethics and the Ada Lovelace Institute, was conceived to help address these fundamental questions. By pulling together existing research and expertise on the current state and expected trajectory of AI-powered genomic science, and undertaking an explicit analysis of the power dynamics likely to be created as a result, this project aims to understand how AI-powered genomics may impact people and society and what steps policymakers can take to address these issues.
This two-year project makes use of different research methods and activities to address a series of interconnected questions. The first half of the project, devoted to understanding the current state of AI-powered genomics, involved a literature review, horizon scanning, and quantitative analysis of research, investment and patent activity. The second half of the project (to be published in 2024) makes use of scenario mapping, public deliberation and policy stress-testing and development to understand the implications of these findings.
This report sets out what we have done (and found) in the first phase of the project, and the specific questions and areas we are focusing on in the second phase. It also details the ambitions and scope of the project, the methodology that we have developed to help us deal with a complex, expansive set of questions and the most potentially significant developments at the intersection of AI and genomic science.
In this report, we focus on setting out the areas where investigation and new policy thinking is most needed. While we provide some suggestions as to where the most immediate policy questions concerning AI-powered genomics might lie, we will be developing firmer conclusions and recommendations for policymakers over the next phase of the project, which will be published in 2024.
There are other aspects of genomic science that the project does not cover. As we detail more fully in the next chapter, our work is concerned with the nature and impacts of observational genomics technologies, rather than interventional ones. As such, we are interested in AI-powered genomic analysis, as applied to the human genome, but do not explore the ramifications of AI-powered advances in gene editing technologies such as CRISPR-CAS-9 (for example). Likewise, the project is focused on how genomic analysis might be used to improve understanding of human biology and medicine, and therefore will not directly consider how such advances might yield better insights into pathogens, and non-human animals.
Scope, research questions and methodology
The aim of this report is to provide policymakers and those working on AI-powered genomics with an analysis of the nature and significance of recent and predicted future advances, the factors and actors driving these advances, and the opportunities and challenges posed.
This chapter describes our approach and methodology for answering these questions, setting out the three phases of the project and their research and analysis activities.
The scope of the project
AI and genomics futures is concerned with the societal implications of:
- AI and genomics together, rather than separately
- how AI is influencing genomics – rather than how genomics is influencing AI.
- how AI is influencing genomic analysis – and not genome editing.
- the AI-powered analysis of the human genome and genomes – and not the genomes of diseases, plants and non-human animals
- how AI might influence the analysis of the human genome and genomes over a 5-10 year time horizon
- AI and genomics globally (though in practice some of the more detailed research, exploration and analysis may have a regional focus).
Research questions, phases and activities
| Question | Activity |
| Empirical questions | Research phase |
| How are advances in AI (and in particular machine learning and deep learning) changing the capabilities, viable applications and practice of genomic science? | Literature review, scientometric analysis and horizon-scanning exercises (to survey the trends and likely AI-driven developments in human genomic analysis over the next 5-10 years, and existing debates concerning their potential societal, legal and ethical impacts).
|
| Of these changes, which are most likely to come about and which are most likely to be realised in the short-to-medium term?
|
Prioritisation exercise to select the trends and developments most likely to come to fruition and be societally and politically significant – and therefore worthy of further exploration.
|
| Exploratory/futures-focused questions | Exploration phase |
| What are the potential societal and political economy consequences of these developments? | Scenario-mapping exercise setting out some possible futures that might result from different policy reactions to the emergence of these identified capabilities.
|
| What outside factors might influence the impacts that these changes have on society and the political economy?
|
|
| Normative and policy-focused questions | Development phase |
| What are public priorities and values when it comes to the spectrum of possibilities posed by AI-powered advances in genomic science? | Public deliberation aimed at investigating how different possible futures align with or deviate from public values and priorities, and what the public would want policymakers to do to shape the development of these new genomics capabilities.
|
| What should decision-makers do now and in the future to ensure that predicted and possible developments work for and are in the interests of people and society? | Policy development work to establish whether the public’s recommendations could be transformed into an agenda for policy and regulatory change, and actions to close any identified gaps between the current policy trajectory and such an agenda. |
How we work with others and draw on expertise
The highly technical nature of the subject matter requires us to draw on external expertise and guidance at various stages of the project.
One source of external expertise is the AI and genomics futures Advisory Board, a panel of seven external experts from a diverse range of backgrounds, ranging from genomics and medicine, health economics and law, to futures thinking and bioethics (see below). The role of the Advisory Board is to provide advice and guidance to the project team on the structure, delivery and approach of the project, and to feedback on workshops, planning and documentation.
In addition to this, the project will draw on external expertise in the more substantive questions posed by the different components of the project.
The AI and genomics futures Advisory Board
Rachel Adams
Principal Researcher
Research ICT Africa
Joan Costa-i-Font
Professor in Health Economics
London School of Economics
Sarah Ennis
Professor of Genomics
University of Southampton
Sasha Henriques
Principal Genetic Counsellor
Guy’s and St Thomas’s NHS Foundation Trust
Shwetha Ramachandrappa
Consultant Clinical Geneticist
Guy’s and St Thomas’s NHS Foundation Trust
Laurie Smith
Head of Foresight Research
Nesta
Sheetal Soni
Senior lecturer in Bioethics, International Law and Intellectual Property Law
University of KwaZulu-Natal
The components and methodology of the project
AI and genomics futures is divided up into three main phases.
The research phase
The research phase, which generated the findings set out and discussed in this report, ran from the spring to the autumn of 2022, and aimed at providing a clear overview of the current state and anticipated trajectory of AI-powered genomic analysis, and the associated legal, academic and policy discourse. This phase of the project involved:
- A literature review, conducted over spring and summer 2022 by Arianna Manzini (independent researcher) and Tim Lee of the University of Edinburgh. This focused on how AI is being applied and is hoped to be applied to genomic science, and the current ethical, legal and policy debates concerning AI-powered genomics.
- A scientometric analysis, carried out by the data science team at Nesta (the UK’s innovation agency) over summer and autumn 2022. This took a quantitative look at academic and start-up company databases, patent data, and public and private research funding. The team at Nesta collected data about research, technology development and business activity in the technology and life sciences sectors. Natural language processing (NLP) and machine-learning methods were used to identify emerging trends and themes at the intersection of AI and genomics from the past decade.
The scientometric analysis was aimed at providing a data-driven understanding of trends in academic and industry research. It also aimed to provide insight into current and anticipated business models applying AI-powered genomics, and to identify the most significant public and private funders of research and development – along with the biggest recipients of this investment.
- A horizon-scanning exercise, which used a form of the Delphi Method to ask a panel of 13 external experts, from academia, industry, medical science, Government and consultancy, to put forward their predictions about the most likely, impactful developments in AI-powered genomic science over the next 5-10 years. This process involved:
- a brainstorming phase, in which each participant listed the ten most significant advances in genomic science enabled by the application of AI likely to emerge over the next 5-10 years
- a prioritisation phase, in which each participant reviewed the aggregated, anonymised responses to the brainstorming exercise, and identified the ten most important advances
- a ranking phase, in which participants were presented with the ten advances identified as most important, and scored each for its likelihood of being realised over the next 5-10 years, and its potential scientific or technical significance.
The horizon-scanning exercise generated a clear picture of what experts think to be the most significant and probable developments in the field over the next 5-10 years (as well as generating useful information about how different groups working at the intersection of AI and genomics view the immediate future and significance of the technology).
Contributors to the AI and genomics futures horizon-scanning exercise
Francisco Azuaje
Director of Bioinformatics
Genomics England
Anthony Cox
Principal Scientist
Illumina
Alastair Denniston
Honorary Senior Lecturer
University of Birmingham
Tania Dottorini
Associate Professor in Bioinformatics
University of Nottingham
Faisal M. Fadlelmola
Principal Investigator, Centre for Bioinformatics & Systems Biology
University of Khartoum
Jennifer Harris
Director of Research Policy
The Association of the British Pharmaceutical Industry
Daniela Hensen
Senior Portfolio Manager for Artificial Intelligence
Biotechnology and Biological Sciences Research Council
Priya Kalia
Global Communications
Eagle Genomics
Alex Mitchell
Director of Bioinformatics
Eagle Genomics
Rakhi Rajani
Chief Digital and Strategy Officer
Genomics England
Sobia Raza
Senior Programme Manager at the Big Data Institute
University of Oxford
Doctor Alessandro Riccombeni
National Genomics Officer
Microsoft
Sven Sewitz
Director of Biodata Innovation
Eagle Genomics
William Spooner
Head of Data Infrastructure
FL86
Virginie Uhlmann
Research Group Leader
European Bioinformatics Institute
Topic prioritisation exercise
The final task of the research phase was to collate the topics and themes identified in the literature review, scientometric analysis and horizon-scanning exercise, and to consult with the Advisory Board on which of these should be prioritised for further investigation.
Our objective was to narrow the diverse set of topics identified to a single emerging theme or set of capabilities that is 1) likely to be realised and deployed over the next 5-10 years, and 2) likely to pose challenging, difficult questions for decisionmakers. This exercise was undertaken with the help of our Advisory Board, who scrutinised and helped refine our approach, reasoning and conclusions.
The conclusions of this prioritisation exercise are set out in the ‘Key research findings and their implications’ chapter.
The exploration phase
The principal activity of the exploration phase is a scenario-mapping exercise, focused on mapping out the different ways that the key themes or technological capabilities identified in the research phase might be deployed and impact upon society, the economy and politics.
The purpose of the scenario-mapping exercise is to develop a fuller, more informed picture of how different actors are incentivised and would therefore be likely to behave in the context of the availability of new AI-powered genomic capabilities. The scenario-mapping exercise involved working with our Advisory Board and external partners to describe four possible futures that could emerge within the next 5-10 years as a result of the development and increasing availability of AI-powered genomic analysis.
The development phase
Following identification of the four possible futures of AI-powered genomics , the development phase of the project will focus on the question of how decision-makers can prepare.
The first part of the development phase will be to use public deliberation workshops to discuss what decision-makers should do in response to the potential scenarios that could arise as a result of predicted AI-powered developments in genomics.
Workshop participants will be introduced to the subject matter and background issues, presented with the four futures identified in the exploration phase, and invited to reflect on how Government (and other key decision-makers) might attempt to guide technological developments in light of these possibilities.
The second part of the development phase will assess where and how the recommendations developed by the public might be converted into more concrete policy suggestions or steers for Government. Here, we are likely to deploy a combination of gap analysis and policy research, along with engagement and expert workshops.
Detailed research findings
This chapter sets out the findings from the literature review, the scientometric analysis and the horizon-scanning exercise.
AI-powered developments in genomics: Findings from the literature review
The first section of the literature review provided a clear overview of how AI is contributing to and predicted to contribute to genomic science:
- Upstream contributions, where AI is improving the collection and processing of data and other prerequisites for genomic analysis.
- Core contributions, where AI is improving the analysis and interpretation of genomic data.
- Downstream contributions, where AI is making it more viable to apply and deploy genomic insight in products and services. (For example, with AI-powered chatbots being used by non-specialist clinicians to understand and communicate the implications of genetic tests.)
Upstream contributions
AI is often seen as a tool with the potential to both improve the supply of good quality, representative data needed for genomic analysis – and to enable genomic analysis to be conducted with less and worse quality data than otherwise possible.
On the supply side, machine-learning techniques can help address issues with errors and noise in genomic data obtained through DNA sequencing, thereby improving its accuracy.[18]
Moreover, machine learning techniques like natural language processing (NLP)– a computational technique aimed at analysing and synthesising natural language and speech – are also cited as a means to speed up and reduce the human resources required for the preparation and interpretation of phenotype data (clinical information such as a patient’s disease symptoms, sex, age — which is an important complement to genomic data in much genomic analysis).[19] It has also been suggested that AI might enable medical data (such as doctor’s notes) to be put into a machine-readable format, which makes it easier to study genotype/phenotype correlations across different sources of data. [20] Likewise, AI has the potential to improve the speed of genomic (and phenotypic) data sharing with, for instance, the development of novel algorithms to package whole genomic datasets into smaller parts.[21]
On the demand side, the academic literature identifies AI as a tool capable of lowering some of the currently high resource and data requirements for effective genomic analysis. An important feature of AI is its capacity to enable robust inferences from smaller and lower quality data sets than would otherwise be possible. For instance, AI can be used to make predictions about the value of gaps in a genomic sequence.[22] Applied to genomics, AI therefore has the theoretical capacity to make genomic analysis less ‘data hungry’.[23] [24]
Core contributions
Perhaps most fundamentally, AI is often described in the literature as a powerful tool in overcoming the often prohibitive complexity of genomic data and its interpretation.
Our literature review and horizon-scanning exercise both homed in on several anticipated applications of AI for analysing large quantities of complex data. These include:
- The use of computer vision (a field of AI focused on the recognition of patterns from digital images) for better identification of phenotypic and genetic variations between humans. [25] For instance, computer vision has been applied to microscopic images of lung cancer material to identify cancerous cells, determine their type, and predict genetic variations (specifically somatic mutations) present in a tumor.[26]
- Deploying AI to improve understanding of the non-coding portion of the human genome, [27] which is the 98 percent of the human genome that does not directly code for proteins, but is thought to be responsible for how genes are expressed.[28] One example is the use of a deep-learning technique (deep matrix factorization) to better understand complex relationships between long non-coding RNAs in the expression of human diseases.[29]
- Ambitions to use AI to make medically actionable distinctions between patients on the basis of their genotypes, including better prediction of individual disease risk and of drug responses.[30] (It should be noted that accounts differ as the speed at which medically useful inferences about patient phenotypes might be made from genomic data, with some suggesting that this will be a slow process.)[31] [32]
Within the medical literature, there is a heavy focus on the potential for AI to improve understanding of, and potential treatments for, cancer (as a ‘disease of the genome’).[33] More generally, there is emphasis on the potential for AI-powered genomics to identify genes and gene mutations associated with disease, enabling the development of drugs that target these genes specifically.[34]
Downstream contributions
AI also has potential as a tool for helping non-specialists understand and apply insights from genomic analysis. One example of this is the use of natural language processing (NLP) in helping non-specialist medical professionals to deploy insights from genomic testing in clinical settings. The PHG Foundation’s 2018 report on AI and genomics in health cites the example of the ‘Genetic Information Assistant’ created by Clear Genetics, and GeneFAX by OptraHealth, which are AI-powered chatbots, designed to help explain the implications of genomic analysis to patients, and we continue to see similar tools in use 5 years later.[35] [36]
Current limitations to the application of AI to genomics
One major barrier to AI-powered genomic analysis is a comparative lack of phenotype data (data about patients’ observable physical characteristics). In order to analyse the roles played by specific combinations of genes, it is necessary to assess the correlations and patterns between organisms’ genomes and their phenotypes. Currently, however, far more genomic data is being gathered than usable phenotype data, which tends to be slower and more complicated to collect, as well as being more variable and open to interpretation.[37]
A related caveat concerns AI systems’ ability to predict phenotypic data – such as the likelihood of being diagnosed with a particular disease – on the basis of genomic data. Many frameworks and tools developed to predict particular physical or behavioural attributes on the basis of genomic variations have not been experimentally validated – meaning that it is as yet unclear whether or not they would hold up in real-world settings. While this lack of experimental validation does not necessarily rule out the predictive accuracy of these tools, it does stand in the way of their adoption in clinical settings.[38]
Similarly, the literature review notes that the lack of transparency of machine- and deep-learning systems is a significant obstacle to their adoption in clinical settings, where clinicians typically need to understand the causal reasoning for diagnoses and suggested interventions to be able to act on them.[39] As such, while AI systems are expected to complement human medical expertise, many researchers do not expect that AI systems will lessen the need for the judgements of trained clinicians in the short-to-medium term.[40]
Figure 1: Ways AI could overcome longstanding challenges in genomics – and current limitations
| Challenge in genomic science | Potential contribution of AI | Limitations to AI’s current use |
| The preparation and interpretation of phenotype data is labour intensive | AI could automate and speed up the collection and processing of phenotype data | · There is a comparative lack of phenotype data against which to compare genomes.
· Some AI-powered predictive models lack experimental validation. · – The lack of transparency of some AI systems’ reasoning might prohibit use of AI powered genomic insight in clinical settings. |
| Effective genomic analysis requires large amounts of data | AI could make genomic analysis less data hungry | |
| Genomic data, and data analysis, is exceptionally complex | AI can help identify patterns in genomic data difficult to find by other means | |
| Understanding and applying insights from genomic analysis requires specialist training | AI (natural language processing (NLP) could help make genomic insights understandable to non-specialists |
Legal, ethical and societal challenges
The second part of the literature review identified several distinct issues concerning the legal, ethical and societal ramifications of the application of AI to genomic science.
One set of ethical challenges relates to the limitations of biomedical datasets and AI systems. Datasets required for training and deploying AI-powered genomic analysis systems are prone to several shortcomings.
For example, like other types of biomedical data, genomic and phenotype data is inherently prone to noise and variation, meaning that datasets often contain corrupted, incorrect, or irrelevant data.[41]
Crucially, noise is often created at source – at the point of data collection or generation – making it harder to identify and account for at a later stage. For instance, in medical and healthcare contexts, noise is often a consequence of errors with equipment or techniques used, which lead to results that do not represent the true nature of the biological material being studied.[42] With genomic data, the DNA extraction process is often probabilistic and can therefore add erroneous data. Other reasons for noise include the loss of metadata and the fact that criteria for applying particular categories or labels to data can often be ambiguous, or include difficult edge cases.
Additionally, genomic datasets often represent very particular demographic groups – the majority of data in major genomics data banks, for instance, capture information about people from white European ancestries, and lack information about people from African ancestries.[43] Lastly, historical phenotype data is often labelled in a way that reflects the prejudices of those responsible for the labelling. [44] Clinical notes recorded by physiatrists reflect the historical tendency to make different treatment recommendations for ethnic minority groups and female patients.[45] These inadequacies can lead to AI systems exhibiting poor predictive accuracy, different rates of accuracy for different groups and machine bias – where the system makes incorrect assumptions about connections between different data points.
The literature also discussed the specific difficulties with preserving the privacy of genomic data and insight. Three major concerns are:
- The results of genomic tests have implications for a subject’s relatives. An individual deciding to take a genomic test for a genetic disease, for instance, may inadvertently reveal whether or not a close relative has that disease. In such cases, a person’s interest in understanding their health might conflict with their relatives’ desire to not know about theirs. It also means that people who want to keep their genomic details private may struggle if their relatives are more willing to share the results of genomic tests.
- Future inferences that may be made from an individual’s genomic data cannot be known at the time of sharing. For this reason, it is hard for a genomic data subject to fully predict what information they might be committing to reveal about themselves in the future, by sharing their genomic data now.
- Genomic data is particularly difficult to anonymize.[46] [47] A common technique for preserving the privacy of sensitive personal data, particularly when it is collected in large databases for research purposes, is removing aspects that data that might allow it to be linked back to a specific individual (known as anonymisation). While anonymisation can be difficult to achieve in practice with most forms of personal data, genomic data is particularly difficult, if not practically impossible, to anonymise. By comparison to other forms of personal data, very small amounts of genetic information can be used to uniquely identify an individual. Moreover, because large amounts of genetic information are shared between relatives and ethnic groups, it is possible to narrow down the identity of a person in an anonymised genetic database if you have the genomic data of a relative against which to compare it.[48]
A final challenge discussed was around the explainability and interpretability of AI systems tasked with processing genomic data. In a notable point of overlap with the scientific literature, researchers expressed concern about how the opacity of machine- and deep-learning systems makes it difficult to understand how and why such systems reach conclusions.[49] [50]
The literature also includes a set of debates around the specific uses to which genomic analysis might be put, and the impacts of such uses on particular groups, including a worry about ‘social genomics’. This term describes the use of genomic analysis to predict complex behavioural and non-physical traits associated with life outcomes, including educational attainment and socio-economic status. Social genomics is cited as having the potential to enable far more personalised, predictive approaches in domains such as education, criminal justice and recruitment. There are, however, serious concerns about the accuracy (and epistemic grounding) of such techniques, as well as worries that the availability of such insight could lead to genetic discrimination (a term used to refer to the risk that people may be treated differently because they have, or are perceived to have, certain genetic variants). [51] [52] [53]
There was a group of questions about the cost, and opportunity cost, of investment (and particularly government investment) in AI-powered genomic science, especially in the context of healthcare. Given current shortcomings in access to conventional medicine and care, and the numerous environmental factors contributing to poor population health, there is a debate about whether the considerable investment required to realise the promise of genomic medicine can be justified.
Proponents[54] of investment argue that AI-powered genomic medicine will ultimately pay dividends by enabling more effective medical interventions and more efficient allocation of healthcare resources. It was also argued that some of the high costs associated with investment in genomic analysis, such as the high monetary and energy costs of data collection and storage, may decrease with time as a result of the increasing viability of cloud and distributed computing.[55]
On the other hand, sceptics of investment express unease about substantial spending on a set of technologies whose efficacy is not yet proven, and whose impacts will not be felt for a considerable period of time. In response to predictions of falling costs for genomic data processing, researchers point to the Jevons paradox to suggest that any increases in efficiency will be matched by increases in demand, thereby negating any overall cost savings.[56]
Sceptics of investment in AI-powered genomics also point out that any benefits that do arise from genomic medicine are likely to be unevenly distributed in the absence of more substantial changes to healthcare provision at a global level.[57] There are also concerns about the vision of AI-powered genomics as a tool to inform resource allocation, with a particular concern about systems distributing resources on the basis of overall system efficiency, rather than on the basis of individual need.[58] [59]
Finally, a comparatively small part of the literature surveyed focused on how the development of and availability of genomic analysis might influence, and be influenced by, existing economic and political power dynamics, and in particular, the relationship between state, citizen and the private sector. One of the most common concerns expressed in the academic literature was about how access to and control over the data and processing capacity required to conduct genomic analysis is likely to be concentrated in the hands of a small number of private-sector companies operating in Europe, North America and East Asia. Researchers contended that this dynamic could make it difficult for governments and non-state actors to regulate, steer and deploy AI-powered genomics in the public interest, and could lead to research and development priorities tailored predominately to the concerns of rich nations.[60] [61]
Emergent ethical and societal questions
It is striking that very few of the ethical issues identified in the academic literature are unique to the application of AI to genomics specifically. The majority of the debate focuses on ethical and legal issues common to genomics, genetics and other applications of AI predictive analytics. Indeed, the list of concerns around privacy, bias, discrimination and opportunity cost are familiar from discussions about AI and genomics when they are considered separately.
This absence doesn’t mean that the emergence of AI-powered genomics presents no new normative questions or challenges. Instead, it is far more likely to be because the convergence of AI and genomics is still a relatively new phenomenon (whose particular ethical implications are simply too novel to have been covered in any detail).
In mapping out the legal, ethical and societal challenges posed specifically by AI-powered genomics, it is useful to distinguish between those that are variations on familiar normative questions, and those that have no clear analogue from existing debates.
The former are challenges where the character of existing concerns about AI or genomics look substantially less or more acute where the two technologies are combined. For instance, it may be that, by making genomic analysis faster and easier to apply, AI increases the number of instances in which we may potentially run into ethical problems posed by that analysis. Similarly, it may be that genomics constitutes an especially sensitive type of data for AI to be applied to, making concerns about AI bias and discrimination far more vivid and consequential than in other contexts.
It may also be that existing concerns about broader, societal and political consequences of genomic insight only become serious should AI lead to faster, cheaper genomic analysis than is currently available.
Challenges of the latter kind (in which the application of AI to genomic science leads to ethical or societal challenges that are not currently posed by AI or genomics alone) are harder to map out. Totally new ethical challenges will either come from completely novel technological capabilities (which are hard to predict) or they come from the emergence of new economic, political or societal dynamics.
Some potential candidates for novel challenges of this kind relate to the merging of the cultures and incentives of technology companies and the life sciences, or the consequences of healthcare becoming de facto dependent on sensitive personal data (which raises questions about privacy, consent and the healthcare quid pro quo).
Despite being hard to predict, these novel challenges are important to try and think about because they can potentially present the biggest unexpected problems for policymakers.
A key objective of the exploration phase of this project, and in particular, our use of scenario mapping and public deliberation will be to tease out some of these novel dynamics and understand how they might present challenges for policymakers and other decision-makers.
Meta trends in the application of AI to genomics: Findings from the scientometric analysis
Our commissioned scientometric analysis provides useful insight into where AI-powered genomics is being researched, by who, and with what focus.
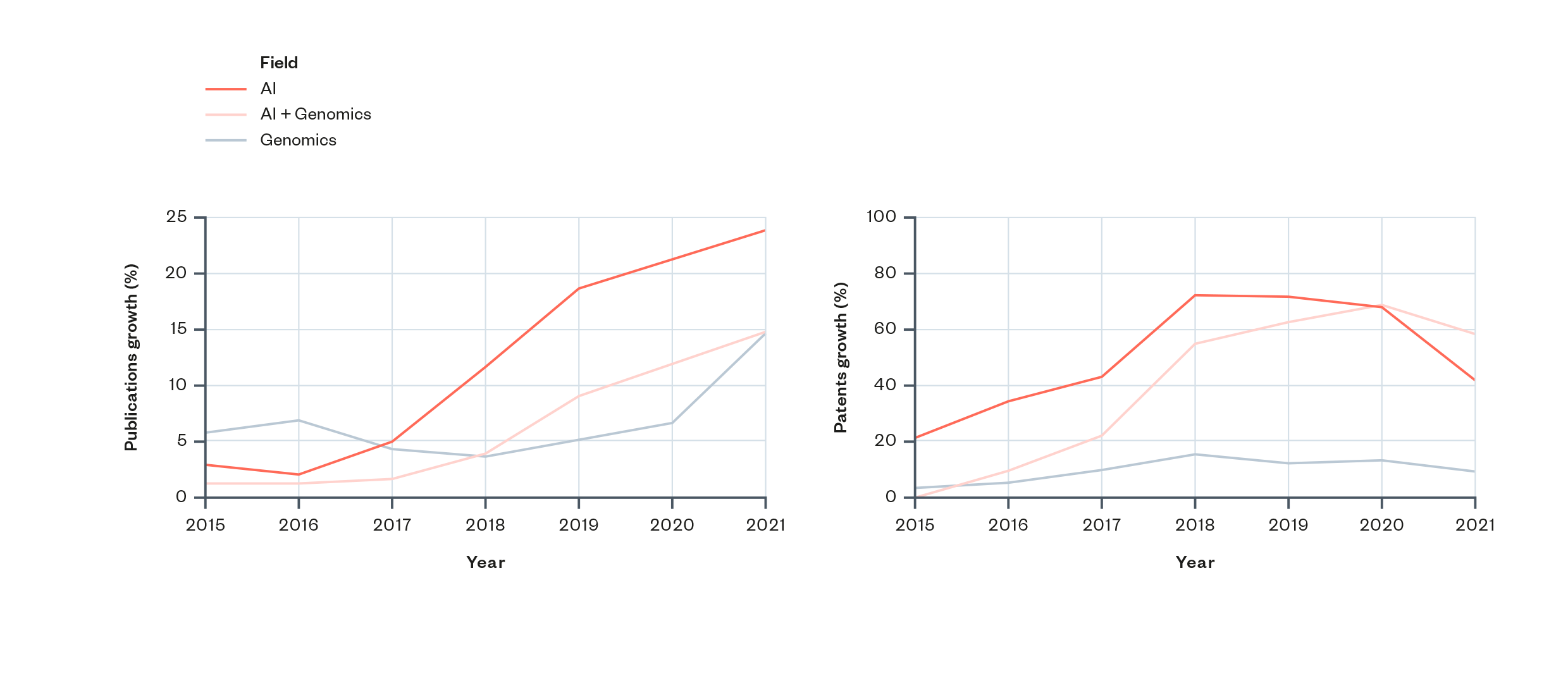
- Overall levels of research and patenting in AI and genomics have been rising since the mid-2010s. There is a particularly pronounced and continued rise in the rate of academic research in this area (see Figure 2).
- The timing supports the idea that growth in AI and genomics has been driven by the rise and increasing prominence of deep learning and artificial neural networks within the field of AI. The increase in levels of research activity in AI and genomics coincides with the permeation of deep learning and neural networks throughout the field of machine learning. This could suggest that the uptake of these methods (or the ability to perform them), has driven an increase in AI and genomics. (More detail is provided in the scientometric analysis section ‘Disciplinary influence and influence of research’.)
- Research in AI and genomics has become more advanced, with new clusters of topics (such as around data compression, graph analysis and neural networks) emerging over the past decade.
- Research has also become more specialised, with an increasing number of research topics unique to the intersection of AI and genomics having emerged over this time period. Moreover, there has been a shift in the research from a small number of relatively broad topic clusters to a larger number of narrower topics.
Figure 2: Increases in publication and patenting activity in AI and genomics since 2015[62]
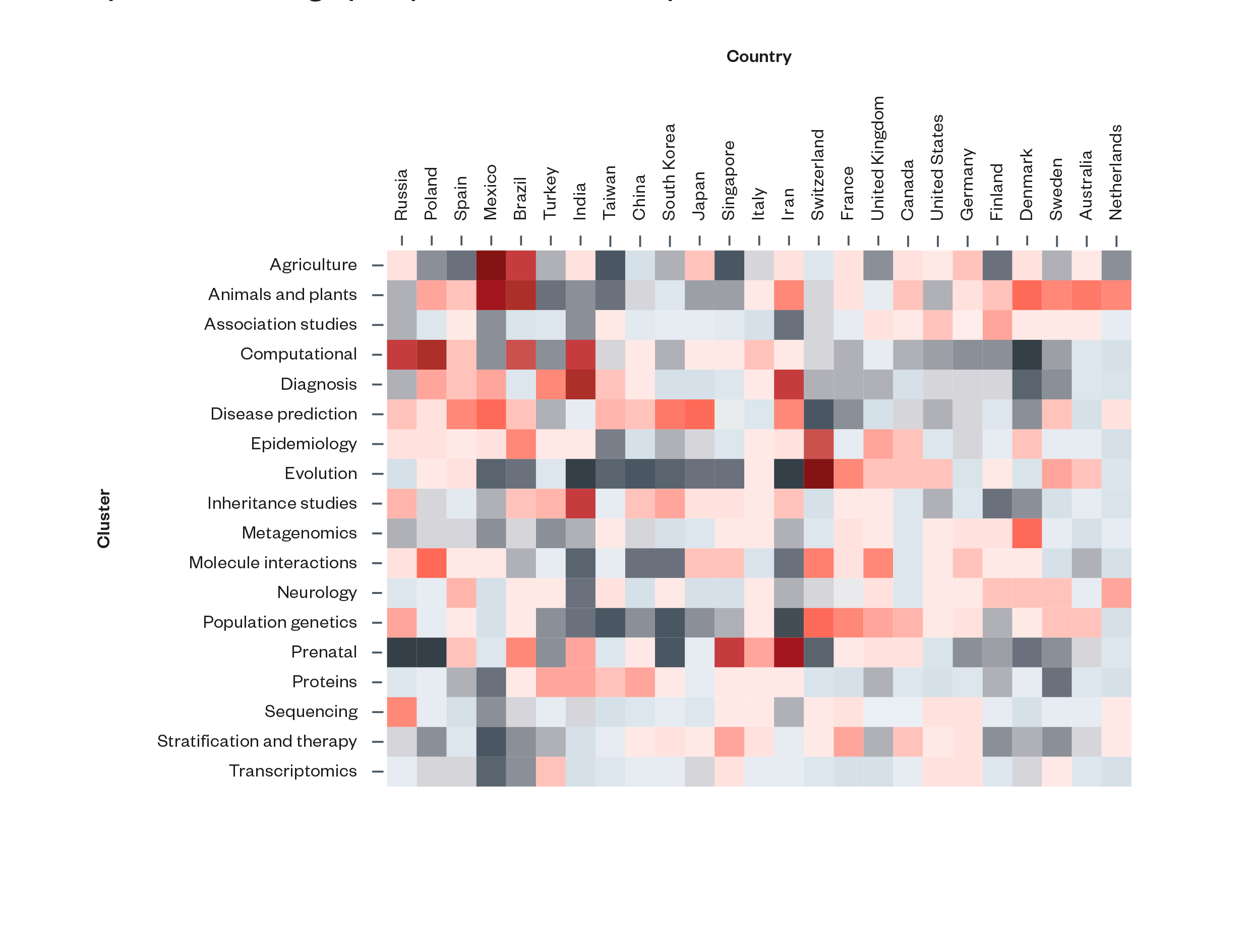
There is a broad geographical divide in academic specialisation on AI and genomics. The scientometric analysis revealed some notable differences in research specialisation between different countries and regions of the world. Countries in Asia tend to have a greater degree of specialisation in disease prediction and proteins, and tend to be less specialised in population genetics. Likewise, countries in North America and Northern Europe tend to have greater specialisations in association studies and population genetics.
It is also notable that very few (8 percent) of the countries surveyed showed a degree of specialisation in metagenomics, with the highest being Denmark. The most activity in that topic comes from the Technical University of Denmark.
Figure 3: Distribution of specialisation in AI and genomics topics across countries by publication contributions. Redder squares represent higher areas of specialisation and greyer squares lower levels of specialisation[63]
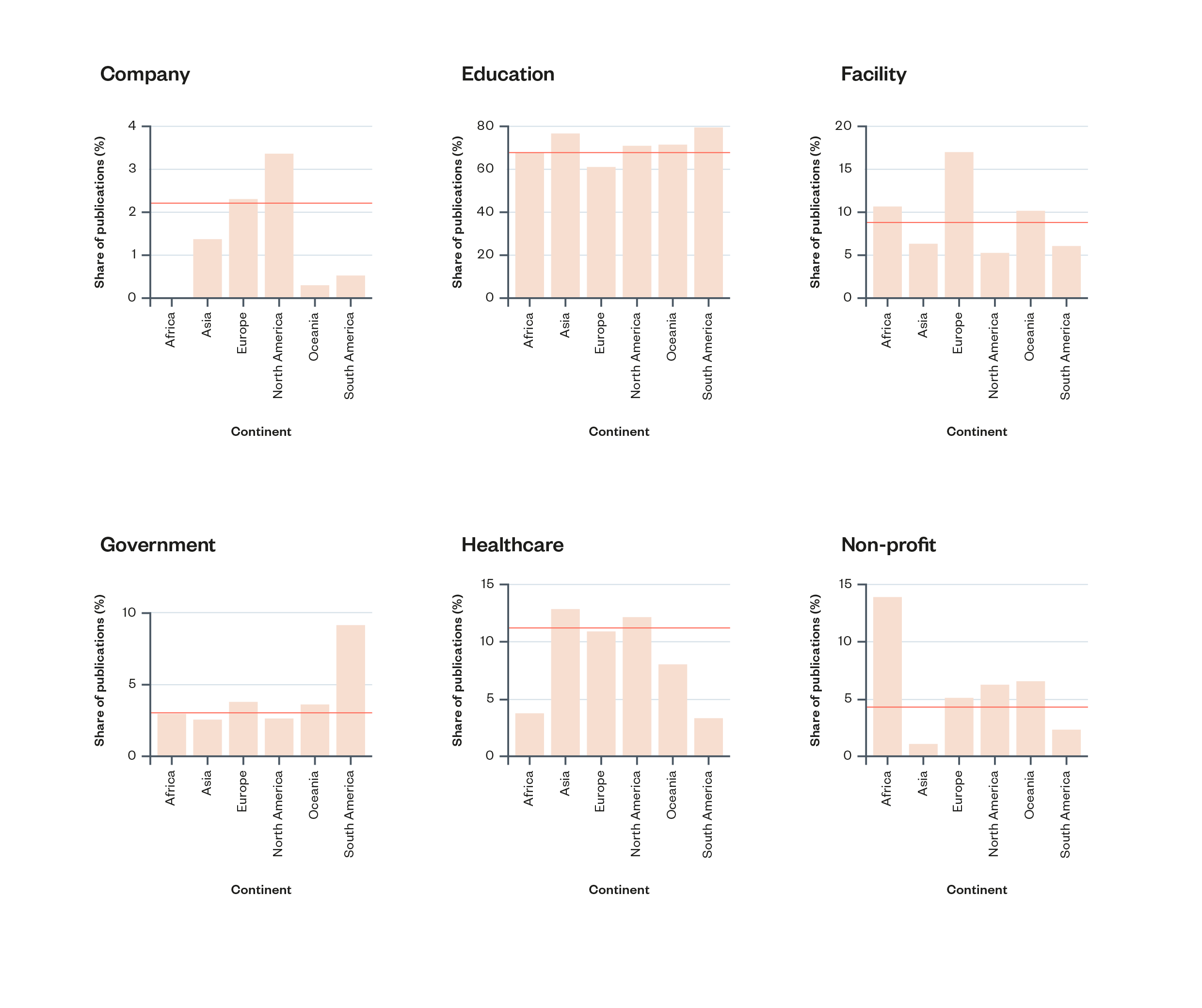
There are some notable differences between the kinds of institutions working on AI and genomics in different regions of the world. In Europe and (especially) in North America, companies are responsible for a higher proportion of academic publications than in other regions. In South America, government institutions make up a higher proportion of research activity. Africa has the highest proportion of research by non-profits of any region (though this is accounted for almost entirely by one institution: Cape Town HVTN Immunology Laboratory).
Figure 4: Percentage of AI and genomics publication contributions by institution type within each continent. Yellow horizontal lines show each institution type’s global share of contributions[64]
Within countries, academic research and commercial R&D on AI and genomics tend to focus on different topics.
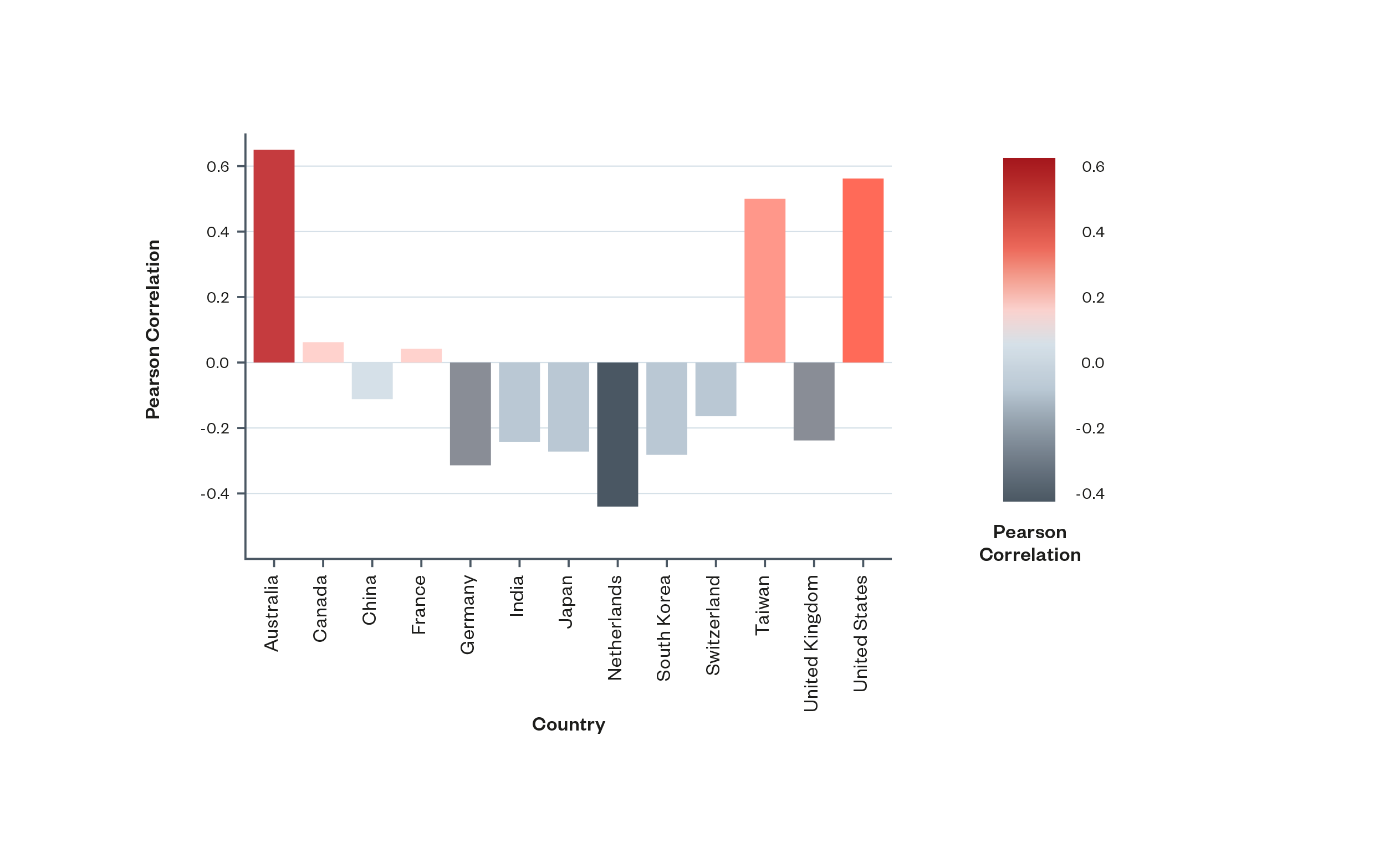
By comparing rates of specialisation across academic databases and patents, the scientometric analysis found that within most countries, commercial R&D tended to focus on different areas to academic research. Notable exceptions to this trend were the United States, Australia and Taiwan, each of which showed positive correlations between commercial and academic specialisations.
There are a few different possible explanations for this phenomenon, including: academic researchers and companies are pursuing different priorities; research in AI and genomics does not typically translate into commercial application within national borders; and companies draw on an international (rather than domestic) base of research when developing products and services.
Figure 5: Pearson Correlation[65] of the distribution of topic specialisation in publication and patenting activities within countries. Most countries studied demonstrate negative or weak correlations between specialisation in academic research and patents, with Australia, Taiwan and the United States notable exceptions[66]
Universities, healthcare institutions and government research institutions tend to focus on different aspects of AI and genomics.
The scientometric analysis found that education institutions (universities and similar entities) have the most highly diversified publication activity. Healthcare organisations, by contrast, tend to exhibit a much narrower research focus, with strong specialisations in a handful of clinically relevant areas, such as association studies, disease prediction and stratification and therapy. Government institutions appear to specialise in areas that are sensitive to national security (and that are outside the scope of AI and genomics futures) such as agriculture, animals and plans and epidemiology.
Figure 6: Institution types and publication activity. Redder squares indicate higher levels of publication activity and greyer squares indicate lower levels[67]
Trends and predictions for the next 5-10 years: Common themes from the horizon scanning and scientometric analysis
On the question of which specific advances in AI-powered genomics are most likely to be realised over the course of the next five to ten years, the findings of the horizon-scanning exercise and the scientometric analysis are well aligned, with some common, overlapping themes identified across the two pieces of research.
Drug development and personalised medicine are the topics that come across most strongly from both the horizon scanning and the scientometric analysis.
- Drug development (the use of AI to combine genomic data with clinical, biological and phenotype data to speed up the discovery of new drug targets and the search for compounds capable of affecting identified targets) was identified by the horizon-scanning panel as both highly probable and highly impactful. The scientometric analysis also shows that companies working on drug discovery are amongst those current attracting the most funding and (alongside those working on target identification) are amongst those who have raised the most money to date.
- Personalised medicine (the use of an individual’s genomic data to guide decisions about the detection, prevention and treatment of disease) was identified by the horizon-scanning panel as relatively likely and highly impactful. (‘Precision medicine’, as it was referred to in the horizon-scanning exercise, scored 5.6 for likelihood and 8.4 for impact.) In the scientometric analysis, it is a topic that falls under both the ‘stratification and therapy’ and ‘association studies’ concept clusters, which were both assessed to be highly significant in terms of overall research and patent activity.
Multiomic analysis and polygenic analysis were topics that came through strongly in the horizon-scanning exercise, and which had a relatively high degree of representation in the scientometric analysis.
- Multiomic analysis[68] was identified by the horizon-scanning panel as both highly probable and highly impactful (with scores of 8.6 and 7.6 respectively). While it did not come through clearly as a subtopic of any of the research clusters in the scientometric analysis, the analysis did show that companies working on the combination on genomic and non-genomic datasets are amongst those currently attracting the most funding.
- Polygenic analysis[69] that individually have small impacts, and which is cited as a means to investigate the genomic basis of complex traits) was identified in the horizon scanning as one of the most likely developments of those considered (with a score of 7.1), and to be relatively high impact (with a score of 6). Though it does not come up prominently as a concept in its own right in the scientometric analysis, it accounts for 11 percent of the papers under the highly significant ‘association studies’ topic cluster.
A final mention should be given to transcriptomics/non-coding variant analysis,[70] which featured clearly, though not as prominently, in both the scientometric analysis and the horizon-scanning exercise. The horizon-scanning panel were not strongly convinced that transcriptomics was a development likely to be realised within the next 5–10 years (giving it a likelihood score of 5.8). This assessment echoes the findings of the scientometric analysis, which identified transcriptomics as well represented and fast growing within the academic literature, but not yet a significant presence in the patent databases (which tend to pick up advances that are more mature, and therefore closer to practical or commercial application).
The direction of funding and private-sector activity within AI and genomics
The scientometric analysis revealed that, of organisations working on AI and genomics:
- Those working in precision medicine, drug discovery or building integrated genomics AI platforms are raising the most amount of money over more funding rounds.
- In terms of total funding, companies related to data collection specifically, drug discovery, precision medicine and target identification have raised the most funding to date.
- Many of those who have been through the highest total number of funding rounds to date (a measure that indicates the maturity of organisations) are organisations working on topics such as genomics AI platforms, cancer and biomarker discovery.
Figure 7 : A plot of the results of the horizon-scanning process. Topics deemed most probable are plotted furthest to the right, and those assessed to be most impactful are plotted closest to the top.
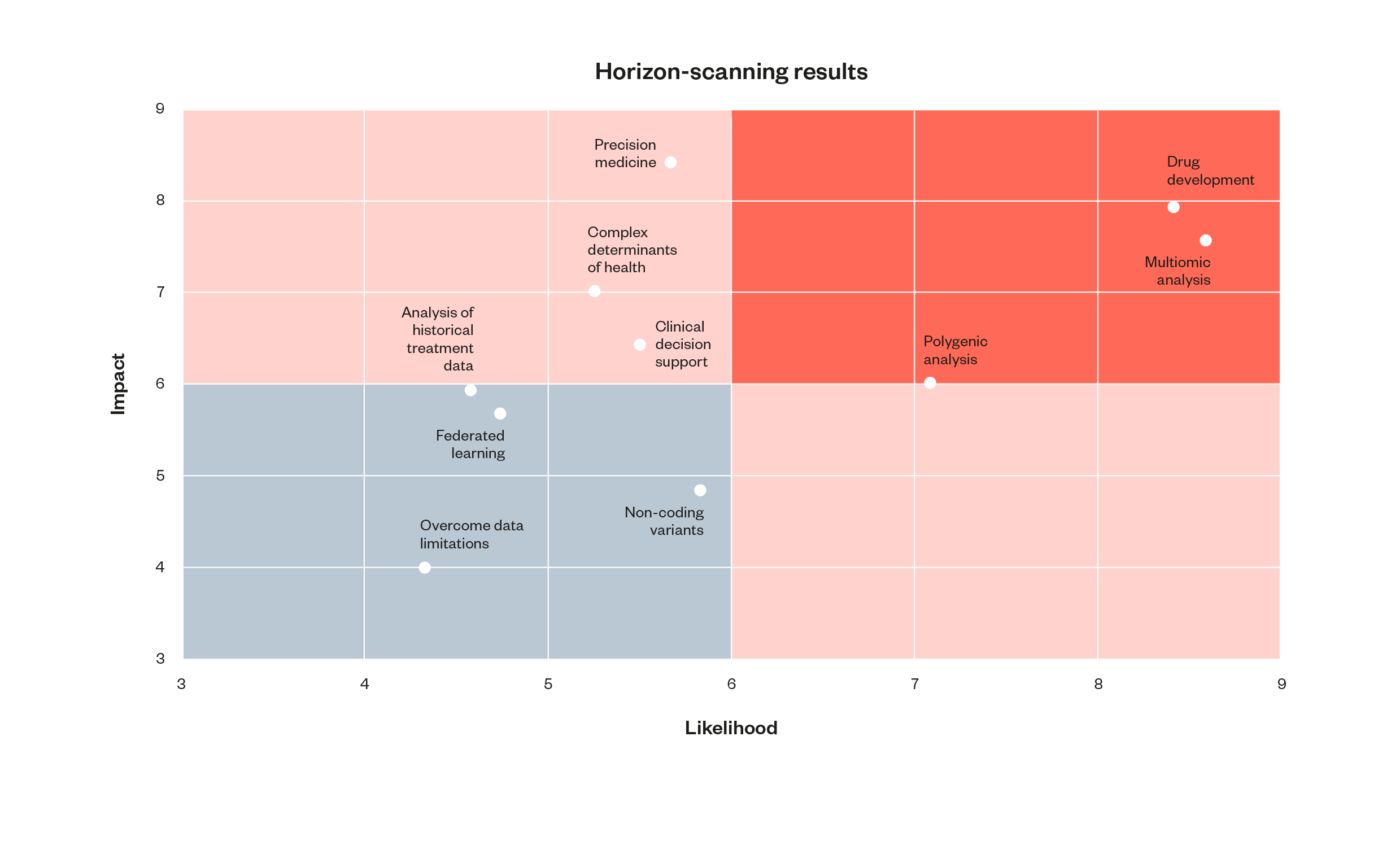
Figure 8: A table of the results of the horizon-scanning exercise
This table provides further detail on each of these horizon-scanning topics, along with more precise figures on the panellists’ assessments.
The figures in the likelihood column denote the mean average of horizon scanning panellists’ scoring of each topic out of ten, in terms of their assessment of the ‘likelihood of being substantially realised and deployed within the next 5-10 years’.
The figures in the impact column denote the mean average of panellists scoring of each topic out of ten, in terms of their assessment of ‘the significance of its realisation and/or deployment from a scientific or clinical perspective’.
| Predicted advance | Description/explanation | Likelihood | Impact |
| AI enables analysis that combines genomic data with many other datasets (multiomic analysis) | Multiomics performs analysis across combined data modalities (omes) including genomics. Such analyses are enabled by machine learning as they are often, following appropriate pre-processing and feature selection, agnostic to the data modality. These analyses typically result in predictions of multi-factor associations between molecular features and disease-related variables. | 8.6
|
7.6
|
| AI enables faster drug development | AI enables the combination of genomic data with associated clinical, biological and phenotype data to greatly speed up the discovery of new drug targets and the search for compounds capable of affecting identified drug targets. | 8.4
|
7.9
|
| AI enables more sophisticated polygenic analysis | Polygenic analysis estimates the effect of many genetic variants on an individual’s phenotypes (observable traits). Whereas traditional methods are limited to the fitting capabilities of the linear model, machine-learning methods introduce non-linear models and capture interaction information between single variants. | 7.1
|
6
|
| AI enables better assessment of the molecular consequences of non-coding variants | 90 percent of the genetic variants associated with disease do not directly alter proteins. Most of these relationships are empirical (e.g. from GWAS studies) but the application of AI techniques can help to unpick their molecular basis, especially in combination with other data modalities (omes) such as 3D structure of the genome. | 5.8
|
4.8
|
| AI enables the effective practice of precision medicine | The application of AI improves 1) the accuracy and viability of pharmacogenomics, and 2) the speed and practicality of genome sequencing, enabling drug prescriptions and treatment decisions to take account of genetic variation. | 5.6
|
8.4
|
| AI is deployed to support clinical decision-making informed by genomics | AI ‘clinical decision support systems’ (powered by natural language processing (NLP) systems) enable non-specialist clinicians to apply genomic insight in clinical settings. | 5.5
|
6.4
|
| AI enables better understanding of complex (genomic and non-genomic) determinants of health | The use of AI to integrate and find patterns in complex ‘-omics’, healthcare, environment, lifestyle, biosocial and other datasets could be used to generate novel insights. These could be used to produce better understanding of health inequalities, to establish biomarkers of health and predict resilience to health conditions across the lifespan. | 5.3
|
7
|
| AI enables federated learning | AI enables federated learning, whereby genomic (and non-genomic) data from multiple databases could be mined for novel insights. | 4.8
|
5.6 |
| AI enables the efficient analysis of historical treatment data | The application AI powered text recognition and NLP enables medical data to be efficiently collated and interpreted, and used in genomic research. | 4.6
|
5.9
|
| AI is used to overcome limitations of small or fragmented genomic datasets | Examples of AI being used to overcome small or fragmented genomic datasets include:
· potential strategies for synthetic data to complement ‘real world’ health data in clinical trials whilst maintaining public trust · the use of AI to parameterise interpretable models allowing mechanistic inference · the use of AI to uncover complex interactions within very large-scale fragmentary data such as those arising from microbiome research and rare disease research. |
4.3
|
4
|
Figure 9 : Summary of findings of the scientometric analysis[71]
This table provides a ranking of the most prominent topic clusters identified by the scientometric analysis. ‘Hot’ topics are those where levels of patent and academic research activity are both high and have been growing recently. ‘Stab’ (or stabilising) topics are those where levels of patent or academic research activity are high, but where recent activity is lower or has slowed recently. The level of private-sector participation for each topic is measured by the number of academic papers in that topic identified as having at least one private-sector contributor.
| Topic | Definition | Academic status |
Patent status | Topic(s) or titles of 3 most prominent papers | Level of private-sector participation |
| Stratification and therapy | Identification of population segments to predict patient risk and treatment outcomes | Hot | Hot | Cancer prediction; cancer diagnosis; cancer personalised treatment | Low |
| Proteins | Analysis and prediction of protein structures and sequences | Hot | Hot | Protein folding; MicroRNAs and complex diseases; genome annotation | Low |
| Association studies | Association studies, including Genome Wide Association Studies (GWAS) | Stab | Stab | Whole-genome regression for quantitative and binary traits; Pharmacogenomics Knowledge for Personalized Medicine; Precision medicine in 2030—seven ways to transform healthcare | High |
| Sequencing | Sequencing methods and studies | Stab | Stab | Single-cell RNA-seq preprocessing; Next-generation sequencing technologies, Efficient assembly of nanopore reads | High |
| Diagnosis | Machine learning methods applied to diagnostic methods and imaging | // | Hot | Cell type discrimination in single cell analyses; Gene selection | Medium |
| Transcriptomics | RNA artifacts, sequencing and selection | Hot | // | Spatial transcriptomics, prioritization and exploratory visualization of biological functions; scMC learns biological variation through the alignment of multiple single-cell genomics datasets | Medium |
| Inheritance studies | Trait inheritance | // | Stab | Enhancer grammar in development, evolution, and disease: dependencies and interplay; An efficient medical image encryption using hybrid DNA computing and chaos in transform domain; Evaluation of extreme precipitation over Asia in CMIP6 models | Low |
| Metagenomics | Big data and machine learning methods applied to metagenomics | // | Stab | Microbiome analysis; detection of diverse DNA and RNA viruses; Quantitative image analysis of microbial communities with BiofilmQ | High |
Key research findings and their implications
Over the course of 2022, the AI and genomics futures project team conducted a series of research activities to better understand:
- How AI is changing and predicted to change the capabilities and viable applications of genomic science.
- Which emerging and predicted changes are most likely to be realised and widely exploited over the next five to ten years.
A key aim of this research was to identify a specific application of AI-powered genomics that is 1) likely to occur in the future and 2) could have significant impacts on people and society.
The second phase of the project will use scenario mapping to explore the consequences of this application in more detail, and public deliberation to assess preferences and views on how it should be managed.
Our research revealed:
In the short-to-medium term, the vast majority of applications of AI-powered genomics are likely to be in medical settings. Of the research, development and business activity identified by our research, almost all was in or was most directly relevant to healthcare or medicine.[72]
The topics that emerged the most prominently, and those on which significant progress was deemed most probable within the next 5-10 years, were:
| Emergent theme | Significance/application |
| Proteins | Drug discovery and development |
| Association studies | Understanding of the population-level correlation between a genetic variant and a given trait |
| Stratification and therapy | Prediction of patient and group disease risk, and personalisation of treatments based on genotype |
| Polygenic analysis and scoring | Diagnosis and prediction of complex traits |
| Multiomics | Analysis combining genomic and non-genomic datasets |
| Pharmacogenomics | The prediction of drug responses and drug response variation between different genotypes |
Considered together, these developments suggest that, over the coming decade, AI-powered genomics has the potential to contribute to and accelerate the technical viability of two broad practices within medical and healthcare settings:
1. Genomic personalisation: The ability to understand how treatment needs for the same condition might vary between different individuals or groups, and to tailor and adapt treatments accordingly.
AI-powered improvements to scientific understanding of the relationship between genes and the structures of proteins, and in the diagnosis of particular diseases and disorders, could enable the development of new and bespoke medicines, and the adaptation of these to specific genotypes or disease instances.
2. Genomic prediction: The use of data to estimate the probability of different individuals or groups developing particular conditions, their responses to particular medicines or treatments, or to predict how their health might be affected by lifestyle factors such as smoking and diet.
AI-driven advances in polygenic analysis and risk scoring, and multiomics could lead to improved insight into the disease risk of different individuals and groups, and how such risks are affected by outside factors. Likewise, advances in pharmacogenomics could enable predictions of patients’ reaction to different medicines or treatments on the basis of their genotype. This information could be used to improve prevention, to enable better planning and more efficient resource allocation.
| Broad capability | Constituent capabilities | Underlying technological advances | |
| Personalisation | Developing new drugs
Tailoring drugs to genotypes |
Proteins, association studies, stratification and therapy | Pharmaco
genomics |
| Prediction | Predicting disease and disease risk
Predicting responses to drugs and environmental stimuli Predicting healthcare needs |
Polygenic analysis and risk scoring, multiomics, | |
The availability of techniques of genomic personalisation and prediction could have significant impacts for both healthcare and wider society – and therefore poses difficult, value-laden questions for decision-makers.
Of these two potential developments, genomic health prediction could pose especially difficult questions for policymakers and healthcare professionals (see the text box below). In particular, these groups will need to address:
- How genomic health predictions can be made and acted on responsibly, in individual cases.
- The impact of the availability of genomic health predictions on the way healthcare is structured, and on broader society.
- The wider impact of the infrastructure, systems and norms developed to enable and exploit the availability of genomic health prediction.
Our focus on AI-powered genomic health prediction
After narrowing down to these two technology areas, our Advisory Board determined that the questions presented by AI-powered genomic prediction are deeper, and more varied than those presented by AI-powered genomic personalisation.
One of the reasons for this difference is that the use of genomic prediction could be harder to detect and monitor than genomic personalisation. Genomic personalisation, which involves medical treatment being tailored to the needs of individual patients, is most naturally deployed as a supplement to existing models of medical care – and would most likely happen in the context of existing patient clinical interactions (or at the point of care). This de facto confinement to clinical settings means that it should be comparatively easy to enforce existing norms about patient consent and awareness. By contrast, genomic prediction does not, in of itself, involve the provision of medical treatment. As a result, prediction could be carried out ‘at a distance’, with subjects having little to no awareness of the fact, and limited ability to consent.
Another relevant difference between personalisation and prediction is the way the two capabilities might be used. Personalised medicine is most likely to be treated as a luxury, capable of improving treatment outcomes (at an additional cost – at least in the short term), because it is tied to the provision of healthcare interventions. Genomic prediction, by contrast, could be marketed as a means to more efficiently allocate existing, finite healthcare resources, or as a means to target public health measures aimed at prevention and demand reduction.
While existing legal, ethical and policy debates provide some guidance on how some aspects of these questions might be navigated, there are many areas where further thinking is required.
Specifically, there is a need for deeper, more sustained exploration of the implications of AI-powered genomic health prediction as a subset of AI-powered genomics, and how these might implications be managed. In contrast to the existing discourse, this exploration will need to pay particular attention to:
- The distinct, emergent issues presented by AI-powered genomic health prediction, as well as those that are analogues from AI ethics and from bioethics.
- The political economy of AI-powered health prediction, and how the technology will shape – and be shaped by – political and economic power dynamics, and how different ways of realising the technology will impact these dynamics differently.
This is thinking that will require consideration of the different, concrete material conditions in which genomic health prediction might develop, and without recourse to the normative judgements of members of the public.
Emergent ethical and political issues presented by genomic health prediction will only become apparent after the fact, or at the very least, when the specific circumstances of the technology’s realisation are modelled.
Likewise, questions of how policymakers should attempt to direct genomic health prediction cannot be disentangled from broader discussions about the kinds of societal relations and dynamics that are and aren’t desirable – in other words, about the kind of world we all want to live in. Answers to such heavily value-laden questions must be informed by the public.[73]
The focus of our research moving forward
The exploration phase of AI and genomics futures will involve a deep dive into societal and political implications of anticipated advances in genomic health prediction over the next 5–10 years, and the levers and options available to policymakers.
In order to better map out the potential issues posed by genomic health prediction, we will use scenario mapping to articulate some of the ways this technological capability could manifest and be deployed, given uncertain background conditions.
We will then engage with experts and the public on how genomic health prediction might be managed and directed, given these different possibilities. This insight will be used to develop recommendations for policymakers.
Acknowledgements
This report was authored by Harry Farmer, with substantial input from Andrew Strait and Francine Bennett (Ada Lovelace Institute), and Catherine Joynson and Peter Mills (Nuffield Council on Bioethics).
About the Nuffield Council on Bioethics
Developments in biomedicine and health are essential to solving the world’s problems but can also raise profound ethical challenges. The Nuffield Council on Bioethics (NCOB) was established by the Nuffield Foundation in 1991 to help address those challenges and ensure changes in biomedicine and health benefit everyone equitably and fairly. Since 1994, we have been co-funded by the Nuffield Foundation, Wellcome, and the Medical Research Council.
The NCOB is a leading independent policy and research centre, and the foremost bioethics body in the UK. We are made up of a team of Council members and Executive staff who identify, analyse, and advise on ethical issues in biomedicine and health so that decisions in these areas benefit people and society.
Through our horizon-scanning programme, we monitor bioscientific and medical developments that raise ethical questions and could have impacts on society. We aim to anticipate these developments at an early stage, so that we can consider them and make appropriate recommendations in a timely way.
For over thirty years, we have identified and tackled some of the most complex and controversial issues facing societies across the globe. We have brought clarity to complexity and plotted practical ways through seemingly intractable dilemmas. This has led to shifts in public understanding and lasting policy change in the UK and internationally.
Find out more:
Website: nuffieldbioethics.org
Twitter: @Nuffbioethics
Email: bioethics@nuffieldbioethics.org
Footnotes
[1] GOV.UK. ‘The UK Science and Technology Framework’. Accessed 2 August 2023. https://www.gov.uk/government/publications/uk-science-and-technology-framework/the-uk-science-and-technology-framework.
[2] GOV.UK. ‘UK Completes over 2 Million SARS-CoV-2 Whole Genome Sequences’. Accessed 2 August 2023. https://www.gov.uk/government/news/uk-completes-over-2-million-sars-cov-2-whole-genome-sequences.
[3] ‘AlphaFold’. Accessed 2 August 2023. https://www.deepmind.com/research/highlighted-research/alphafold.
[4] Shinwari, Zabta Khan, Faouzia Tanveer, and Ali Talha Khalil. ‘Ethical Issues Regarding CRISPR Mediated Genome Editing’. Current Issues in Molecular Biology, 2018, 103–10. https://doi.org/10.21775/cimb.026.103.
[5] Jones, Elliot. ‘Explainer: What Is a Foundation Model?’ Ada Lovelace Institute, July 2023. https://www.adalovelaceinstitute.org/resource/foundation-models-explainer/.
[6] Such as the DeepMind’s development of Enformer, an AI tool that has led to improvements in predicting how a gene in a DNA sequence might be expressed: Avsec, Ž., Agarwal, V., Visentin, D. et al. Effective gene expression prediction from sequence by integrating long-range interactions. Nat Methods 18, 1196–1203 (2021). https://doi.org/10.1038/s41592-021-01252-x
[7] One example of this is a surge of interest in applying AI techniques to genomic data for drug discovery: Eisenstein, M. Machine learning powers biobank-driven drug discovery. Nat Biotechnol 40, 1303–1305 (2022). https://doi.org/10.1038/s41587-022-01457-1
[8] P&S Intelligence. ‘AI in Genomics Market Outlook | Revenue Estimation Report, 2022-2030’. Accessed 2 August 2023. https://www.psmarketresearch.com/market-analysis/ai-genomics-market.
[9] Raza, Sobia. ‘Artificial Intelligence for Genomic Medicine’. PHG Foundation, March 2020. https://phgfoundation.org/media/77/download/artifical-intelligence-for-genomic-medicine.pdf?v=1&inline=1.
[10] India Kerle and others. ‘AI and genomics futures: A scientometric analysis of research and technology development in the intersection of AI and genomics’ (Nesta, 2023) <https://osf.io/24dea>
[11] Some estimates range from $5.72 billion by 2027 (https://www.arizton.com/market-reports/artificial-intelligence-in-genomics-market) and $9,9 billion by 2031 (https://www.alliedmarketresearch.com/ai-in-genomics-market-A11556) to (£19.5 billion by 2030, up from half a billion in 2021 https://www.psmarketresearch.com/market-analysis/ai-genomics-market)
[12] Vuksanaj, Kathy. ‘Expectations for AI in Healthcare Become More Modest’. GEN – Genetic Engineering and Biotechnology News, 4 March 2021. https://www.genengnews.com/insights/expectations-for-ai-in-healthcare-become-more-modest/.
[13] Raza, Sobia. ‘Genomics and Artificial Intelligence – a Good Match?’ PHG Foundation. Accessed 2 August 2023. https://www.phgfoundation.org/blog/genomics-and-artificial-intelligence.
[14] Challen R, Denny J, Pitt M, et alArtificial intelligence, bias and clinical safetyBMJ Quality & Safety 2019;28:231-237.
[15] Meredith Whittaker. 2021. The steep cost of capture. interactions 28, 6 (November – December 2021), 50–55. https://doi.org/10.1145/3488666
[16] Murdoch B. Privacy and artificial intelligence: challenges for protecting health information in a new era. BMC Med Ethics. 2021 Sep 15;22(1):122. doi: 10.1186/s12910-021-00687-3. PMID: 34525993; PMCID: PMC8442400.
[17] Munn, L. The uselessness of AI ethics. AI Ethics (2022). https://doi.org/10.1007/s43681-022-00209-w
[18] https://phgfoundation.org/media/77/download/artifical-intelligence-for-genomic-medicine.pdf?v=1&inline=1
[19] Z. Zeng, Y. Deng, X. Li, T. Naumann, and Y. Luo. 2019. “Natural Language Processing for EHR-Based Computational Phenotyping.” IEEE/ACM Transactions on Computational Biology and Bioinformatics 16 (1): 139–53. https://doi.org/10.1109/TCBB.2018.2849968.
[20] Dias, Raquel, and Ali Torkamani. 2019. “Artificial Intelligence in Clinical and Genomic Diagnostics.” Genome Medicine 11 (1): 70. https://doi.org/10.1186/s13073-019-0689-8.
[21] M. Aledhari, M. D. Pierro, M. Hefeida, and F. Saeed. 2021. “A Deep Learning-Based Data Minimization Algorithm for Fast and Secure Transfer of Big Genomic Datasets.” IEEE Transactions on Big Data 7 (2): 271–84. https://doi.org/10.1109/TBDATA.2018.2805687.
[22] Dias, R., Torkamani, A. Artificial intelligence in clinical and genomic diagnostics. Genome Med 11, 70 (2019). https://doi.org/10.1186/s13073-019-0689-8
[23] Shah, Pratik, Francis Kendall, Sean Khozin, Ryan Goosen, Jianying Hu, Jason Laramie, Michael Ringel, and Nicholas Schork. 2019. “Artificial Intelligence and Machine Learning in Clinical Development: A Translational Perspective.” NPJ Digital Medicine 2: 69. https://doi.org/10.1038/s41746-019-0148-3.
[24] Altae-Tran, Han, Bharath Ramsundar, Aneesh S. Pappu, and Vijay Pande. 2017. “Low Data Drug Discovery with One-Shot Learning.” ACS Central Science 3 (4): 283–93. https://doi.org/10.1021/acscentsci.6b00367.
[25] Mobadersany, Pooya, Safoora Yousefi, Mohamed Amgad, David A. Gutman, Jill S. Barnholtz-Sloan, José E. Velázquez Vega, Daniel J. Brat, and Lee A. D. Cooper. 2018. “Predicting Cancer Outcomes from Histology and Genomics Using Convolutional Networks.” Proceedings of the National Academy of Sciences of the United States of America 115 (13): E2970–79. https://doi.org/10.1073/pnas.1717139115.
[26] Dias, R., Torkamani, A. Artificial intelligence in clinical and genomic diagnostics. Genome Med 11, 70 (2019). https://doi.org/10.1186/s13073-019-0689-8
[27] M. Zeng, C. Lu, Z. Fei, F. -X. Wu, Y. Li, J. Wang, and M. Li. 2021. “DMFLDA: A Deep Learning Framework for Predicting LncRNA–Disease Associations.” IEEE/ACM Transactions on Computational Biology and Bioinformatics 18 (6): 2353–63. https://doi.org/10.1109/TCBB.2020.2983958.
[28] https://www.genomicseducation.hee.nhs.uk/genotes/knowledge-hub/non-coding-dna/
[29] M. Zeng, C. Lu, Z. Fei, F. -X. Wu, Y. Li, J. Wang, and M. Li. 2021. “DMFLDA: A Deep Learning Framework for Predicting LncRNA–Disease Associations.” IEEE/ACM Transactions on Computational Biology and Bioinformatics 18 (6): 2353–63. https://doi.org/10.1109/TCBB.2020.2983958.
[30] Ching, T, DS Himmelstein, BK Beaulieu-Jones, AA Kalinin, BT Do, GP Way, E Ferrero, et al. 2018. “Opportunities and Obstacles for Deep Learning in Biology and Medicine.” JOURNAL OF THE ROYAL SOCIETY INTERFACE 15 (141). https://doi.org/10.1098/rsif.2017.0387.
[31] Lefteris Koumakis, Deep learning models in genomics; are we there yet?, Computational and Structural Biotechnology Journal, Volume 18, 2020, Pages 1466-1473, ISSN 2001-0370, https://doi.org/10.1016/j.csbj.2020.06.017
[32] Torkamani A, Wineinger NE, Topol EJ. The personal and clinical utility of polygenic risk scores. Nat Rev Genet. 2018 Sep;19(9):581-590. doi: 10.1038/s41576-018-0018-x. PMID: 29789686.
[33] Vatansever, Sezen, Avner Schlessinger, Daniel Wacker, H. Ümit Kaniskan, Jian Jin, Ming-Ming Zhou, and Bin Zhang. 2021. “Artificial Intelligence and Machine Learning-Aided Drug Discovery in Central Nervous System Diseases: State-of-the-Arts and Future Directions.” Medicinal Research Reviews 41 (3): 1427–73. https://doi.org/10.1002/med.21764.
[34] For example, in: Zampieri, Guido, Supreeta Vijayakumar, Elisabeth Yaneske, and Claudio Angione. 2019. “Machine and Deep Learning Meet Genome-Scale Metabolic Modeling.” PLoS Computational Biology 15 (7): e1007084. https://doi.org/10.1371/journal.pcbi.1007084.
[35] https://phgfoundation.org/media/77/download/artifical-intelligence-for-genomic-medicine.pdf?v=1&inline=1
[36] A current example of the use of a Chatbot for communitating the results of genetic tests is: https://www.invitae.com/en/providers/gia-chatbot
[37] Dias, Raquel, and Ali Torkamani. 2019. “Artificial Intelligence in Clinical and Genomic Diagnostics.” Genome Medicine 11 (1): 70. https://doi.org/10.1186/s13073-019-0689-8.
[38] The conclusion of the Literature Review states that: “The increased number of publications in more recent years indicates that AI and ML research in genomics is expected to continue to rapidly increase. However, one major challenge is that, though many frameworks and tools have been developed to predict various attributes based on genomic data, these require experimental validation in order to be considered for clinical translation.”
[39] For instance: Radakovich, Nathan, Matthew Cortese, and Aziz Nazha. 2020. “Acute Myeloid Leukemia and Artificial Intelligence, Algorithms and New Scores.” Best Practice & Research. Clinical Haematology 33 (3): 101192. https://doi.org/10.1016/j.beha.2020.101192.
[40] Jessica Morley, Caio C.V. Machado, Christopher Burr, Josh Cowls, Indra Joshi, Mariarosaria Taddeo, Luciano Floridi, The ethics of AI in health care: A mapping review, Social Science & Medicine, Volume 260, 2020, 113172, ISSN 0277-9536, https://doi.org/10.1016/j.socscimed.2020.113172.
[41] Ching, T, DS Himmelstein, BK Beaulieu-Jones, AA Kalinin, BT Do, GP Way, E Ferrero, et al. 2018. “Opportunities and Obstacles for Deep Learning in Biology and Medicine.” JOURNAL OF THE ROYAL SOCIETY INTERFACE 15 (141). https://doi.org/10.1098/rsif.2017.0387.
[42] Caudai, Claudia, Antonella Galizia, Filippo Geraci, Loredana Le Pera, Veronica Morea, Emanuele Salerno, Allegra Via, and Teresa Colombo. 2021. “AI Applications in Functional Genomics.” Computational and Structural Biotechnology Journal 19: 5762–90. https://doi.org/10.1016/j.csbj.2021.10.009.
[43] Kessler, M., Yerges-Armstrong, L., Taub, M. et al. Challenges and disparities in the application of personalized genomic medicine to populations with African ancestry. Nat Commun 7, 12521 (2016). https://doi.org/10.1038/ncomms12521
[44] Suresh, Harini, and John V. Guttag. 2021. “A Framework for Understanding Sources of Harm throughout the Machine Learning Life Cycle.” In Equity and Access in Algorithms, Mechanisms, and Optimization, 1–9. https://doi.org/10.1145/3465416.3483305.
[45] Chen, Irene Y., Peter Szolovits, and Marzyeh Ghassemi. 2019. “Can AI Help Reduce Disparities in General Medical and Mental Health Care?” AMA Journal of Ethics 21 (2): E167-179. https://doi.org/10.1001/amajethics.2019.167.
[46] Caudai, Claudia, Antonella Galizia, Filippo Geraci, Loredana Le Pera, Veronica Morea, Emanuele Salerno, Allegra Via, and Teresa Colombo. 2021. “AI Applications in Functional Genomics.” Computational and Structural Biotechnology Journal 19: 5762–90. https://doi.org/10.1016/j.csbj.2021.10.009.
[47] Azencott, C.-A. 2018. “Machine Learning and Genomics: Precision Medicine versus Patient Privacy.” Philosophical Transactions of the Royal Society A: Mathematical, Physical and Engineering Sciences 376 (2128): 20170350. https://doi.org/10.1098/rsta.2017.0350.
[48] Dankar FK, Ptitsyn A, Dankar SK. The development of large-scale de-identified biomedical databases in the age of genomics-principles and challenges. Hum Genomics. 2018 Apr 10;12(1):19. doi: 10.1186/s40246-018-0147-5. PMID: 29636096; PMCID: PMC5894154.
[49] Dias, Raquel, and Ali Torkamani. 2019. “Artificial Intelligence in Clinical and Genomic Diagnostics.” Genome Medicine 11 (1): 70. https://doi.org/10.1186/s13073-019-0689-8.
[50] Caudai, Claudia, Antonella Galizia, Filippo Geraci, Loredana Le Pera, Veronica Morea, Emanuele Salerno, Allegra Via, and Teresa Colombo. 2021. “AI Applications in Functional Genomics.” Computational and Structural Biotechnology Journal 19: 5762–90. https://doi.org/10.1016/j.csbj.2021.10.009.
[51] Comfort, Nathaniel. 2018. “Sociogenomics Is Opening a New Door to Eugenics.” MIT Review Technology, 2018. https://www.technologyreview.com/2018/10/23/139420/sociogenomics-is-opening-a-new-door-to-eugenics/.
[52] Williamson, B. 2020. “Bringing up the Bio-Datafied Child: Scientific and Ethical Controversies over Computational Biology in Education.” ETHICS AND EDUCATION 15 (4): 444–63. https://doi.org/10.1080/17449642.2020.1822631.
[53] Wachbroit, Robert. 2002. “Genetic Determinism, Genetic Reductionism, and Genetic Essentialism.” Encyclopedia of Ethical, Legal and Policy Issues in Biotechnology.
[54] Palmer, Stephen, and James Raftery. 1999. “Opportunity Cost.” BMJ 318 (7197): 1551. https://doi.org/10.1136/bmj.318.7197.1551.
[55] A Jamal, A.R. 2021. “Precision Medicine: Making It Happen for Malaysia.” Malaysian Journal of Medical Sciences 28 (3): 1–4. https://doi.org/10.21315/mjms2021.28.3.1.
[56] Weidinger, Laura, John Mellor, Maribeth Rauh, Conor Griffin, Jonathan Uesato, Po-Sen Huang, Myra Cheng, Mia Glaese, Borja Balle, and Atoosa Kasirzadeh. 2021. “Ethical and Social Risks of Harm from Language Models.” ArXiv Preprint ArXiv:2112.04359.
[57] Hummel, P, and M Braun. 2020. “Just Data? Solidarity and Justice in Data-Driven Medicine.” LIFE SCIENCES SOCIETY AND POLICY 16 (1). https://doi.org/10.1186/s40504-020-00101-7.
[58] Cohen, I. Glenn, Ruben Amarasingham, Anand Shah, Bin Xie, and Bernard Lo. 2014. “The Legal And Ethical Concerns That Arise From Using Complex Predictive Analytics In Health Care.” Health Affairs 33 (7): 1139–47. https://doi.org/10.1377/hlthaff.2014.0048.
[59] Nuffield Council on Bioethics. 2018. “Artificial Intelligence (AI) in Healthcare and Research.” https://www.nuffieldbioethics.org/publications/ai-in-healthcare-and-research.
[60] Lévesque, Maroussia. 2019. “Looking Back to the Future of AI.” Indigenous AI (blog). January 13, 2019. https://www.indigenous-ai.net/looking-back-to-the-future-of-ai.
[61] Stahl, Bernd Carsten. 2021. “Ethical Issues of AI.” Artificial Intelligence for a Better Future: An Ecosystem Perspective on the Ethics of AI and Emerging Digital Technologies, March, 35–53. https://doi.org/10.1007/978-3-030-69978-9_4.
[62] India Kerle and others. ‘AI and genomics futures: A scientometric analysis of research and technology development in the intersection of AI and genomics’ (Nesta, 2023) < https://osf.io/24dea/>
[63] Ibid.
[64] India Kerle and others (n 62)
[65] The Pearson correlation coefficient is the most common way of measuring a linear correlation. It is a number between –1 and 1 that measures the strength and direction of the relationship between two variables. Negative figures denote a negative correlation and positive figures a positive correlation.
[66] India Kerle and others (n 62)
[67]Ibid.
[68] Analysis that combines genomic data with data from other sources – for example other ‘omics’ technologies such as proteomics and epigenomics, and patient medical records and data on environmental factors – to provide a better understanding of the significance of genomic variation.
[69] Analysis looking at the cumulative effect of genetic variations — called single nucleotide polymorphisms (SNPs).
[70]Transcriptomics refers to the analysis of an organism’s whole RNA transcript. RNA is responsible for how the genetic material encoded in DNA is expressed— and therefore affects the relationship between genotype and phenotype.
[71] India Kerle and others (n 62).
[72] Despite being potential uses for genomic analysis often mooted by academics, our research, which covered academic databases, patent activity and public and private research funding, found practically no evidence of scientists or companies openly looking to develop or deploy techniques genomic analysis in areas such as security, education, recruitment or sport.
[73] The context dependent nature of these considerations means that it won’t be enough to rely on the results of previous engagement exercises on similar topics. Though there have been recent dialogues on specific topics relating to genomics (such as those Commissioned by Genomics England on the genomic medicine and the social contract, and on newborn screening), new engagement is required that speaks directly to the above questions.
Related content
AI and genomics futures
This joint project with the Nuffield Council on Bioethics explores how AI is transforming the capabilities and practice of genomic science.

Minding the genomic data gap: COVID-19, genomics and health inequalities
The role of genomics in the data-driven pandemic response